Chromosomal Aneuploidy and PND
Chromosomal anomalies reflect on the average number of chromosomes or a structural abnormality in one or more chromosomes. They usually occur when there is an error in cell division, namely, mitosis and meiosis. The major part of chromosome abnormalities, such as translocations, occurs as an error during the egg or sperm cell division called meiosis. Therefore, they can be inherited from a parent, and in this condition, the abnormality is present in every cell of the body. Some abnormalities, however, are “de novo” manifestations in which the chromosome defects are resulting from mitosis, and thus some cells have the abnormality and some do not. All types of chromosomal abnormalities are categorized into two major groups; numerical and structural anomalies. Numerical abnormalities are aneuploidies defined as either a chromosome from a pair (monosomy) or extra to the normal pair (trisomy). Structural defects are when a part of a chromosome is omitted, additional, substituted to another chromosome, or inverted. Each group of chromosomal anomalies can pose a severe threat to fetus life, indicating the importance of detecting these abnormalities before birth.
Structural abnormalities
Structural chromosomal abnormalities result from inappropriate segregation of chromosomal segments and rejoining in a different gene order after breakage. The structural rearrangements in a chromosome can occur in two main ways; Balanced and unbalanced. Balanced changes happen if the full set of chromosomes is still conserved and only rearranged, including inversion or translocation in chromosomal regions. In an inversion, a segment of a chromosome separated from an arm is turned upside down and reattached to the same place, resulting in the reversed genetic material. However, in translocations, a portion of one chromosome is detached and transferred to another chromosome, leading to two main types of translocation; reciprocal and Robertsonian translocation. Reciprocal changes include the exchange of the segments from two different chromosomes, while in a Robertsonian translocation, a complete chromosomal arm has attached to another at the centromere. Since there is no loss or gain in the DNA material, balanced chromosomal rearrangements may have not any clinical exposure as they do not result in disease. The only condition that results in disease is the breakage that occurs in a gene of each chromosome, leading to missed or nonfunctional protein. Furthermore, if the new chromosome’s combination produces a hybrid of two genes, a new protein will provide a synthesis that harms the cell. However, if the information is added or missing, the rearrangement defines as unbalanced and includes deletions, duplications, or insertions of a chromosomal segment. Deletions (known as partial monosomies) arise in any part of any chromosome when a portion of the chromosome is missing. When there is just one break in the chromosome, the deletion is called a terminal deletion since the end part of the chromosome is missing. Isochromosomes are an example of terminal deletion that can form by a missing arm of the chromosome and duplication of the remaining arm. On the other hand, when a chromosome undergoes two breaks, the deletion is called an interstitial deletion because a piece of chromosome material is lost from within the chromosome. Ring chromosomes are examples of interstitial deletion in which the ends of both arms are deleted, and the remaining sticky ends rejoined to make a circular chromosome. Removals that are too small to be detected under a microscope are called microdeletions. Some of the more common chromosome deletion syndromes include cri-du-chat syndrome and 22q11.2 deletion syndrome. Chromosomal duplication, also known as partial trisomy, occurs in any site of the chromosome as a repeated segment and results in extra genetic material. Some examples of duplication syndromes include 22q11.2 duplication syndrome and MECP2 duplication syndrome.
Numerical abnormalities
Chromosomal abnormality is mostly defined as aneuploidy, a condition that one or more extra copies of a chromosome are present in the cell, owing to a missing or extra copy number of a chromosome. Common fetal aneuploidies include aneuploidies of chromosomes 13, 18, 21, and sex chromosomes 45, X (Turner syndrome); 47, XXY (Klinefelter syndrome); 47, XYY; and 47, XXX. The increased risk of genetic error occurrence has a direct relation to maternal age and several environmental factors. In recent decades, prenatal diagnostic (PND) testing is frequently used for common fetal aneuploidies to detect the abnormal number of chromosomes in the embryo or fetus. Chromosome numerical abnormalities can result in different effects such as pregnancy loss, problems in growth, or congenital disorders depending on the specific copy number changes. According to autosomal trisomies, an additional copy of chromosome 21 causes Down syndrome (trisomy 21), which is manifested by mental retardation, learning difficulties, common facial features, and poor muscle tone (hypotonia) in early stages. Furthermore, in Edward syndrome (trisomy 18), the typical major malformation comprises heart and kidney anomalies. Only half of the babies born with this syndrome live longer than one week, and up to 10% around one year. Lastly, individuals with Patau syndrome (trisomy 13) recognized by microphthalmia, polydactyl, and a cleft lip, often have heart defects and die in the first days or weeks. Only 5-10% of children with this condition survive up to one year. Regarding the sex chromosomal abnormalities, Turner syndrome (TS or monosomy X) is an example of monosomy in which an individual lacks a chromosome. This syndrome is a genetic disorder that occurs solitary in females who they born with only one X chromosome and are usually shorter than average and unable to have children. Moreover, Klinefelter males manifest a range of symptoms that result from two or more X chromosomes, particularly infertility and small poorly functioning testicles. New lines of research on chromosomal aneuploidy disorders demonstrated that genome editing technology could rescue the fetus’s life by the deletion or silencing of an extra chromosome copy. For instance, the interaction of three copies of the RUNX1, ETS2, and ERG genes on chromosome 21 with somatic GATA1 mutations on the X chromosome reported increasing the risk of leukemia in down syndrome, applications of this technique might reduce or overcome the risk of leukemia.
Down syndrome (47, XX+21/47, XY+21)
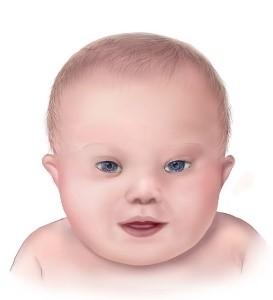
Down syndrome (DS) is a condition that a person has an extra copy of chromosome 21. DS is also referred as to trisomy 21 and it is the most common trisomy in humans. This disease was first described in 1866 by a physician named Langdon Down. The prevalence of DS is 1 in 700-1000 live birth and the Incidence of the down syndrome depends on maternal age. older women are more likely to have babies with Down syndrome. Although Down syndrome is a genetic disorder and related to genes, only 1% of all cases of Down syndrome have a hereditary component (passed from parent to child through genes). DS usually occurs due to the Nondisjunction error during sperm or egg cell division and the formed zygote cell has an extra copy of chromosome 21. during the development of the embryo, the extra chromosome is replicated in every cell of the body. There are three types of Down’s syndrome; Trisomy 21 accounts for 95% of cases, and with this type of DS each cell in the body has 3 separate copies of chromosome 21. Mosaic Down syndrome accounts for 1% of cases, and with this type of DS, there is a mixture of two types of cells, some of them are normal and have 46 chromosomes, and some contain 47 chromosomes with an extra chromosome 21. Translocation Down syndrome occurs in 4% of cases, and in this type, the total number of chromosomes remains 46, but a full chromosome 21 or a part of it is connected to other chromosomes (usually chromosome 14), which causes the symptoms of Down syndrome.
An extra chromosome 21 causes mental and physical disabilities in people with Down syndrome. Affected people have a lower IQ compared to the normal state. Some common physical symptoms of DS include:
- A flattened face
- Almond-shaped eyes that slant up
- A short neck
- Small head and ears
- Shorter in height as children and adults
- Poor muscle tone
There is no cure for Down syndrome but there’s a wide variety of support and educational programs that can help affected people and their families such as speech therapy, occupational therapy, and physical therapy to improve their physical and intellectual abilities.

Edward’s syndrome (47, XX+18/47, XY+18)
Edward’s syndrome or trisomy 18 is the most common trisomy after Down syndrome. This disorder was first described in 1960 by John Edward. Edward’s syndrome is caused by an error in cell division during egg or sperm formation, and the presence of an extra chromosome 18 disrupts the normal pattern of cell growth. the prevalence of Edward’s syndrome is 1 in 5000 live births and a significant number of fetuses with this disorder are never born. Only 50% of affected fetuses survive and they are kept in the intensive care unit after birth. However, many surviving infants also die in the first days of life. Edward’s syndrome is generally more common in girls and girls have more survival chances than boys. Edward’s syndrome includes three types; full trisomy 18, in which all cells in the body have an extra chromosome 18. Mosaic trisomy 18, in which some cells of the affected person’s body have an extra chromosome 18, and partial trisomy 18, in which there is an extra part of chromosome 18 instead of a complete chromosome that is translocated to another chromosome. Full and Mosaic trisomy 18 occur as random events during the formation of eggs and sperm, so they are not inherited. Partial trisomy 18 can be inherited. people who carry a rearrangement of genetic material between chromosome 18 and another chromosome, are at an increased risk of having children with Edward’s syndrome.
Some clinical symptoms of this syndrome include:
- Low birth weight
- Small head and jaw
- An unusual-looking face and head
- Unusual hands and feet with overlapping fingers and webbed toes
- Problems with feeding, breathing, and hearing.

Patau syndrome (47, XX+13/47, XY+13)
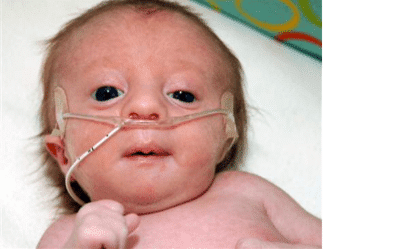
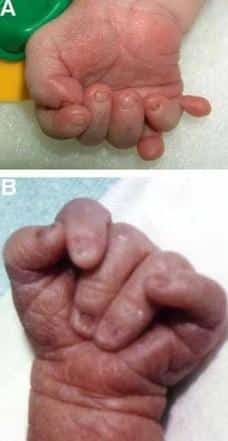
Patau syndrome, which is caused by the presence of an extra chromosome 13, is a very rare disease with an incidence of 1 in 16,000 live births, and its incidence increases with maternal age. Many infants with Patau syndrome die in the first days of their lives. Patau syndrome, like other aneuploidies, is caused by an error in the cell division of the egg or sperm or after fertilization in the zygote cell, and the presence of an extra chromosome 13 leads to brain abnormalities, mental retardation, and muscle and skin abnormalities.
Patau syndrome includes three types, an extra chromosome 13 may be present in all body cells of the affected person (simple or complete trisomy 13) or in some cells (mosaic trisomy 13), and the severity of symptoms is related to the number of cells involved. In some cases, during a translocation between chromosome 13 and another chromosome, the cells resulting from the cell division have an extra part of chromosome 13 (partial trisomy 13). The symptoms and features of both mosaicism and partial trisomy tend to be less severe than in simple trisomy 13, resulting in more babies living longer. Most cases of Patau syndrome occur randomly and are not inherited.
The common clinical symptoms of Patau syndrome include:
- Holoprosencephaly
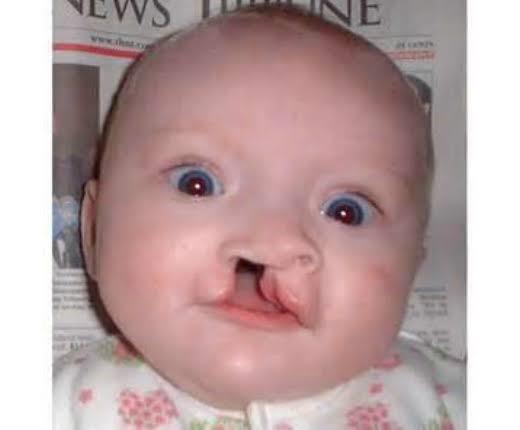
- Facial defects such as the closeness of the eyes, small head and cleft palate, and lip
- Problems with the development of the nasal passage
- An abnormally small eye or eyes (microphthalmia)
- Absence of 1 or both eyes (exophthalmia)
- Ear malformations
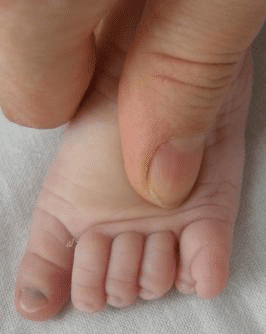
- Extra fingers and toes
- Kidney disorders
- Blindness, deafness, and defects in the scalp
- Abnormalities of the reproductive system

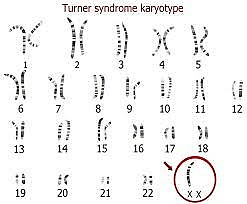
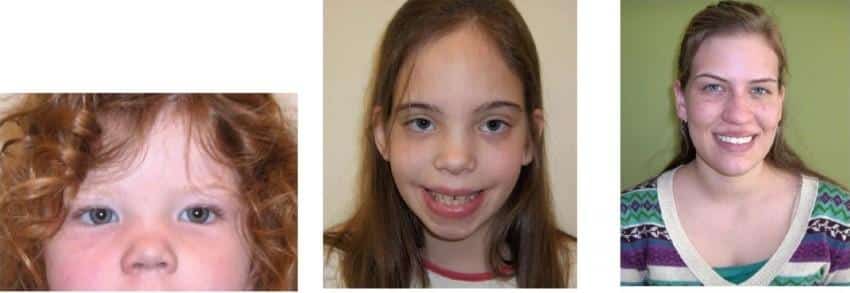
Turner syndrome (46, XO)
Turner syndrome or X chromosome monosomy was first identified by Dr. Henry Turner in 1938 and is the most common genetic disease in girls. Monosomy X is the only monosomy compatible with human life; However, 98% of affected fetuses die spontaneously. In this disease that only affects women, one of the X chromosomes is completely or partially lost. The prevalence of this syndrome is 1 in 2500 live births. This syndrome can occur in two types, classic Turner syndrome (chromosome X completely missing in all body cells) and mosaic Turner syndrome (chromosome X partially or completely missing in some of the body cells). classic Turner syndrome occurs due to nondisjunction error in the sperm or egg formation and mosaic Turner syndrome occurs due to mutations during the early development of the fetus. Turner syndrome occurs randomly, so any types of this syndrome are not inherited. Turner syndrome incidence is not dependent on maternal age.
The genes responsible for the occurrence of Turner’s symptoms in affected people have not been fully identified, but the silox gene, which is involved in bone formation and growth, is one of the identified genes, the loss of this gene probably leads to the shortness of people with Turner’s syndrome.
The common clinical symptoms of this syndrome include:
- Short height (on average 140 cm)
- Infertility – due to underdeveloped ovaries
- Absence of menstruation (amenorrhoea)
- Extra skin (webbing) on the neck
- Hearing problems
- Puffy hands and feet
- Heart and kidney problems
- Crowding of teeth

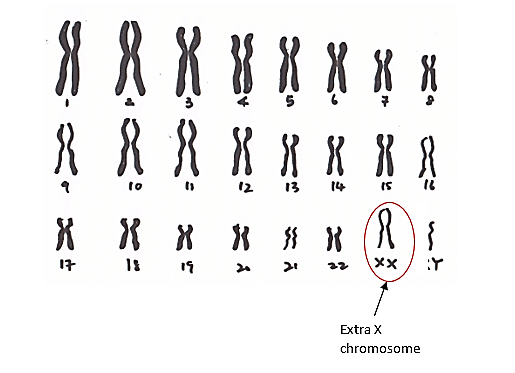
Klinefelter syndrome (47, XXY)
This syndrome is a male-specific genetic disorder in which there is an extra X chromosome in the affected person’s body cells (47, XXY). This disorder is the most sex chromosomal disorder with the prevalence of 1 in 500-1000 live births and is usually not diagnosed until adulthood. This syndrome may be diagnosed when a man realizes his infertility. The incidence of Klinefelter syndrome is dependent on maternal age. An older mother (35 years or older) at the time of pregnancy increases the probability of this syndrome. Klinefelter is caused by an error in fertilization and is not a hereditary disease.
The clinical symptoms of this disease are different in different periods of life, Klinefelter syndrome does not usually cause any obvious symptoms early in childhood, and even the later symptoms may be difficult to spot. Possible features, which are not always present, may include:
- In babies and toddlers: learning to sit up, crawl, walk and talk later than usual, being quieter and more passive than usual
- In childhood: shyness and low self-confidence, problems with reading, writing, spelling, and paying attention, mild dyslexia or dyspraxia, low energy levels, and difficulty socializing or expressing feelings
- In teenagers: growing taller than expected for the family (with long arms and legs), broad hips, poor muscle tone and slower than usual muscle growth, reduced facial and body hair that starts growing later than usual, a small penis and testicles, and enlarged breasts (gynaecomastia)
- In adulthood: infertility and a low sex drive
Jacob’s syndrome (47, XYY)
hypotonia or loss of muscles at birth
Jacob syndrome or superman syndrome is a rare aneuploidy in which men have an extra Y chromosome. The prevalence of this disease is 1 in every 1000 live births. This disorder occurs when the affected person receives an extra Y chromosome with paternal origin and has 47 total chromosomes (XYY). The cause of this problem is the error of Nondisjunction during meiosis and the formation of sexual cells that lead to sperm receiving two chromosomes. An alternate and less common form of this condition is 46, XY/47, XYY mosaicism, which arises during early embryonic development
In some affected men, no symptoms are observed, and the median age of diagnosis is approximately 17 years, with many patients presenting due to infertility concerns. In some cases, there are symptoms include:
- hypotonia or loss of muscles at birth
- tall height
- learning and speaking problems
- The IQ of these people is 10 to 20 units lower than their healthy brother or sister
- These people are also capable of fertility, but in some cases, testicular development problems and sperm or testosterone production defects have been observed.

Triple X syndrome (47, XXX)
Triple X syndrome, XXX syndrome, or superwoman syndrome refers to the presence of an extra X chromosome (XXX) in a girl’s body cells, and the probability of its occurrence is 1 in every 1000 girls born. This syndrome only affects women. The cause of this syndrome is a mistake in the formation of the father’s sperm or the mother’s egg. Sometimes this syndrome occurs during fetal development. In fact, this syndrome is caused by a random genetic error and is not a hereditary disease.
Since in women always one of the X chromosomes is active, many affected people do not have symptoms. But this syndrome can lead to symptoms that include:
- Taller than normal height (without physical problems)
- learning problems
- A decrease of 10 to 20 IQ units and oppositional behaviors
- Early or late puberty
- Small head, upward cleft eyelid, the large distance between the two eyes
- Flat soles
- Low muscle volume
- curvature in the fingers
- Impaired movement and speaking skills

Prenatal Diagnostic (PND) Methods for Aneuploidy Detection
There are different methods for diagnosing Prenatal chromosomal abnormalities. These methods include screening tests and accurate diagnostic tests. Screening tests include NT sonography in the 11-14th week of pregnancy, triple tests (measurement of maternal blood factors) in the 15-18th week of pregnancy, and performing genetic tests using fetal DNA in maternal blood serum (NIPT). Screening tests can be useful in the early stages, but since their results may not be accurate and definitive, more accurate diagnostic methods should be used to be sure. These methods usually require intrauterine sampling such as chorionic villus sampling (CVS) in the 8-12th week of pregnancy and amniocentesis in the 15-20th week of pregnancy. By preparing a karyotype or performing molecular techniques such as FISH, QF-PCR, MLPA, and NGS using DNA extracted from CVS samples, amniotic fluid, and blastocyst, the presence or absence of genetic abnormalities in the fetus can be determined before birth.
Prenatal genetic diagnostic testing is frequently used for common fetal aneuploidies to detect the abnormality of chromosomes in the embryo or fetus. Common fetal aneuploidies include aneuploidies of chromosomes 13, 18, 21, and sex chromosomes (X and Y) that increase sharply with maternal age. Two different diagnostic approaches; namely invasive and noninvasive tests are employed to detect prenatal aneuploidies.
Invasive tests
A. Sampling methods
Invasive diagnostic methods are performed via three biopsy-based procedures such as amniocentesis, chorionic villus sampling (CVS), and percutaneous umbilical cord blood sampling (PUBS). All of these sampling methods pose a risk to mother and fetus and are available only to high-risk pregnancies, which limits the aneuploidy detection rate.
B. Diagnostic methods
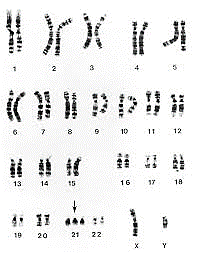
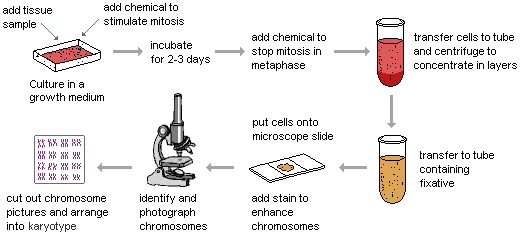
1. Karyotyping
This technique is considered as a gold standard method in which the structure or the number of chromosomes can be detected by a complete picture of whole chromosomes. Karyotyping is a laboratory method that provides the possibility of examining the chromosome set. In this method, after preparing the sample (CVS or the amniotic fluid), the cells in the sample are cultured and then the cells are colored and this color makes the chromosomes visible under the microscope. During this staining, each chromosome is colored with a special pattern of dark and light bands known as G-banding. Usually, GC-rich regions of chromosomes that contain genes show a dark G-banding pattern. Using this method, it is possible to analysis the shape, size, and number of chromosomes. Human chromosome karyotyping means the analysis of 46 human chromosomes in order to detect loss or gain of chromosomes, large deletions, rearrangements (Translocation & Inversion), and duplication. This examination is performed in the metaphase stage of mitosis when chromosomes are in the most intensive state. For this purpose, the cells of the desired sample are multiplied by the cell culture method and collected. Then, with the addition of colcemid (colchicine), cell division stops, and the cells are arrested in the metaphase stage. Metaphase chromosomes are separated, stained, and analyzed using a microscope.
This method is used to diagnose syndromes related to aneuploidy (Down syndrome, Edward syndrome, Patau syndrome, Turner syndrome, etc.). Karyotyping is the gold standard for diagnosing chromosomal abnormalities and detecting common aneuploidies. If a deletion or addition of a chromosome is observed in the child’s karyotype, it is recommended to check the karyotype of the parents for the presence of balanced translocations (which are transmitted to the children).
Different tissues can be used to prepare karyotype, but there is a detection limit for each tissue; For example, a blood sample cannot be used to diagnose trisomy or tetrasomy, and a skin sample is required. Normally, amniotic fluid or chorionic villus are used for prenatal aneuploidy diagnosis.
By using karyotype, deletions and insertion of up to 3-4 million base pairs can be detected. Detection of smaller changes requires molecular cytogenetic methods. Due to the prerequisite of cell culture, the preparation of karyotype usually takes up to 2 weeks. But today, by using molecular techniques, rapid diagnosis is possible in less than 48 hours. The error rate of this method is 0.1 to 0.6%, which is mostly related to the error of gender determination and is due to Maternal Cell Contamination. The time-consuming, labor-intensive and costly nature of this method has increased the tendency to use other methods. However, this method is still used as the gold standard for diagnosing chromosomal abnormalities in diagnostic laboratories.
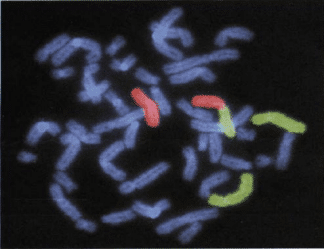
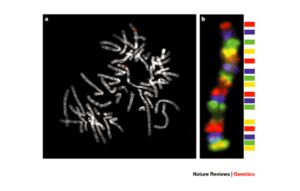
2. Fluorescence in situ hybridization (FISH)
The fluorescence in situ hybridization technique or FISH is another technique used in the diagnosis of aneuploidy. This method can be applied as a fast and relatively specific technique to detect common aneuploidies in samples of uncultured CVS and amniotic fluid. This method combines cytogenetics with molecular genetic technologies and was first introduced by Landegent and his colleagues in 1984. The basis of this technique is the hybridization of a single-stranded DNA called a probe to its complementary sequence in the chromosome. These probes are labeled with fluorescent molecules and fluorochromes molecules allow the hybridized area to be observed using a fluorescent microscope. The chromosome samples that are required in this method can be metaphase and interphase chromosomes, or chromatin strands. In this technique, the probe molecules are directly hybridized into the DNA inside the cell. For this purpose, double-stranded DNA molecules are denatured by increasing the temperature, and the solution containing the probe is added to the sample fixed on the slide. Then, by reducing the temperature, the fluorescent probe molecules hybridize to their complementary sequence in DNA. If the target sequence is present and hybridization is performed, the hybridization position on the chromosome is visible by a fluorescence microscope due to fluorescence emission with a certain wavelength.
There are different types of FISH probes; Centromeric probes are specific for repetitive DNA sequences around or inside the centromere. These probes were initially used to detect common aneuploidies. Specific Unique-Sequence Probes are used to detect deletion or duplication mutations. Whole-Chromosome Paint probes play a role in detecting complex rearrangements.
FISH is used in the diagnosis of cancer, structural abnormalities, chromosome copy number, chromosomal rearrangements, and the diagnosis of infectious diseases. Using this technique, the chromosomal copy number in the interphase nucleus can be determined. In this method, for aneuploidy detection, 24 probes labeled with 5 different fluorescent colors are used. In this way, the examination of 23 autosomal chromosome pairs and 1 sex chromosome pair is possible.
The most important advantage of FISH over conventional karyotyping is no need for cell culture, and results are proved in two days. However, the high cost, the labor-intensive, and the dependence on the user’s skill make this method not generally used in the investigation of chromosomal abnormalities, and it should be performed as a confirmatory method along with other methods.
3. Quantitative fluorescent polymerase chain reaction (QF-PCR)
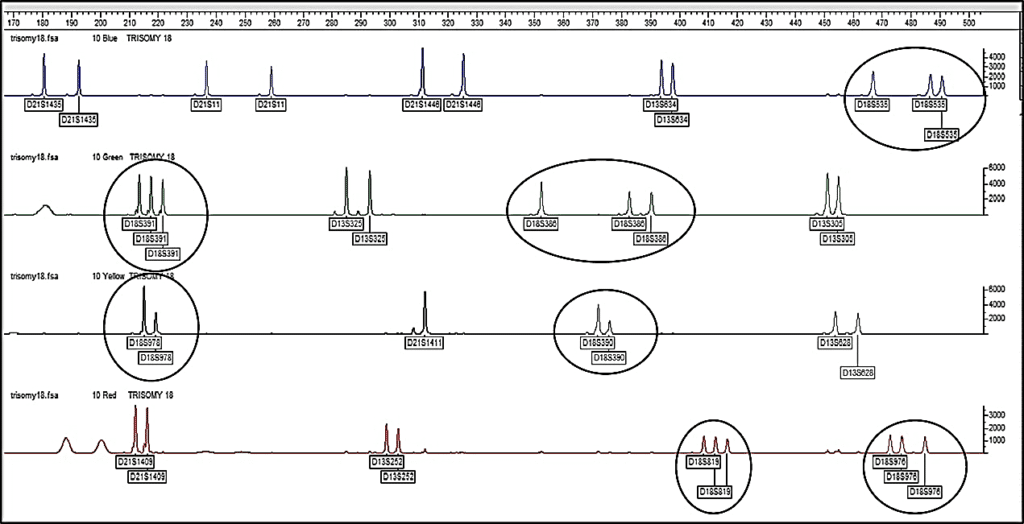
The use of QF-PCR is demonstrated to be a reliable and efficient method for rapid fetal aneuploidy detection. This technique has been used since 1993 for prenatal diagnosis of common aneuploidies (chromosomes 13, 18, 21, X, and Y). It provides a much greater resolution than the conventional karyotyping in combination with the new array CGH method. Furthermore, false-positive results in the NIPT analyses are recommended to be confirmed with the QF-PCR test. This technique uses short tandem repeats, known as STR or microsatellites to identify chromosomal homologs. STRs differ in the number of repeats and the length of the sequence on different loci. Identification and quantification of STRs provide the possibility to determine the chromosome copy number variation.
In this method, after extracting DNA from the samples (amniotic fluid or CVS), amplification of STR markers is done using the PCR method. The primers are labeled with fluorescent molecules, so, after amplification, it is possible to Separation the PCR products by capillary electrophoresis. By interpreting the results of QF-PCR and capillary electrophoresis, it is possible to check the copy number of the examined chromosomes.
The results of capillary electrophoresis are represented as electrophorogram using Gene mapper and Gene marker software. In normal people, where there are two copies of each autosomal chromosome, for heterozygote autosomal markers, two signals with the ratio of 1:1 are created. If a person has trisomy, there are three copies of the corresponding chromosome and two signals are created with ratios of 1:2, 2:1, or three signals of 1:1:1. Therefore, it is possible to detect aneuploidy.
QF-PCR has high accuracy and specificity and its results are prepared within 48 hours. But it should be noted that with this method, it is not possible to detect unbalanced rearrangements and mosaicism of aneuploidies; Therefore, to obtain more accurate results, it is better to perform this molecular technique together with the cytogenetic method (karyotyping).
4. Microarray-based comparative genomic hybridization (array CGH)
This test is a new molecular cytogenetic technique that can look for unbalanced gains or losses of genetic material across the genome. It can find some chromosome problems that karyotyping can miss.
5. multiplex ligation-dependent probe amplification (MLPA)
MLPA is a PCR-based method rely on sequence-specific probe hybridization to genomic DNA that detects copy number changes (like deletions or duplications) of a gene, point mutations, and chromosomal abnormalities. This molecular technique was first introduced in 2002 by Jan Schuten in the scientific journal Nucleic Acid Research. This method is a modified type of the Multiplex PCR method, in which a maximum of 50 gene locus are amplified using only one pair of primers and using special probes. detecting the sequences that only differ in one nucleotide is possible with MLPA.
Performing this technique includes 5 steps; First, the DNA is denatured and the probes (two half-probes) are hybridized with their complementary sequence on the DNA. In the next step, the two halves of the probe are ligated to each other with the ligase enzyme. Then the ligated probes are amplified using fluorescent-labeled primers and finally, the amplified products are separated by capillary electrophoresis and the results are analyzed. A comparison of peaks obtained from the capillary electrophoresis of samples and standard controls, reveals whether the copy number of samples is normal or abnormal.
Using MLPA, it is possible to identify small mutations that the FISH method cannot detect. The efficiency of this method is high and its results are ready within 24 hours. However, this method is not able to detect polyploidy and balanced translocations.

6. Next Generation Sequencing
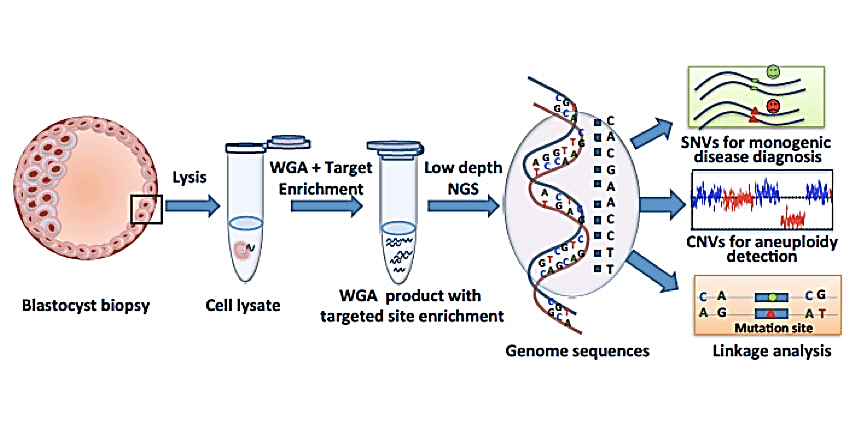
Next Generation Sequencing or NGS is based on the preparation of NGS library in a cell-independent system, where millions of sequencing reactions are performed in parallel and the results are displayed without the need for electrophoresis. In general, in this method, the genomic sample is first divided into a library of small fragments; Then, synthetic oligonucleotides with known sequences are attached to the ends of the fragments; These libraries are then amplified to prepare for sequencing. Today, NGS is used to detect aneuploidy after blastocyst sampling. blastocyst Sampling would allow several cells to be analyzed at the same time and would not include the limitation of scattered sequences that lead to poor diagnosis in PGD.
For chromosome copy number analysis by NGS, the principle involves fragmenting the amplified embryonic DNA into small fragments (100–200 base pairs). Hundreds of thousands of these fragments are sequenced in parallel until a sufficient sequencing depth is acquired. The sequence data from chromosomes across the genome are first compared with the reference genome and then counted with the use of specialist software. Because the number of sequences from a specific chromosome should be proportional to the copy number, trisomy or monosomy will result in greater or lower numbers of reads, respectively.
Using this method and blastomeric biopsy samples, structural chromosomal abnormalities and aneuploidy can be detected. NGS has been validated in different studies with other aneuploidy detection methods and shows 100% accuracy. This method not only detects aneuploidy, but also detects single gene disorders, translocations, and mitochondrial genome disorders. However, one of the disadvantages of NGS is that it is not able to detect unbalanced structural abnormalities.
Non-Invasive Prenatal Testing (NIPT)
A. Sampling methods
Recently, with the emergence of next-generation sequencing technologies, non-invasive prenatal testing (NIPT) has been introduced into clinical practice. In this method, cell-free fetal DNA circulating in maternal blood or locating in the cervical canal is used for aneuploidy assessments. Therefore, the advantage of NIPT is the ability of that in reducing the risk of miscarriage which increases by invasive techniques.
B. Diagnostic methods
1. Fetal cells in maternal circulation
2. cff-DNA in maternal circulation
In these methods, the cell-free fetal (cff) DNA in the maternal circulation is analyzed with direct sequencing of the maternal DNA. They have very high accuracy for detecting abnormal numbers of chromosomes (aneuploidies) (9). There are three main DNA sequencing methods that are used for maternal cff DNA analysis: Massively Parallel Shotgun Sequencing (MPSS), Digital Analysis of Specific Regions (DANSR), and Single Nucleotide Polymorphism (SNP)-based sequencing.
3. Preimplantation genetic diagnosis (PGD)
Preimplantation genetic diagnosis is used for women using in vitro fertilization (IVF) who are at high risk of having a baby with a chromosomal disorder. PGD looks for certain genetic disorders and mutations before an embryo is implemented to the uterus.

